Fill and Sign the Quotation Sample a Form
Helpful hints for preparing your ‘Quotation Sample A’ online
Are you fed up with the inconvenience of managing paperwork? Look no further than airSlate SignNow, the leading eSignature platform for individuals and businesses. Bid farewell to the monotonous task of printing and scanning documents. With airSlate SignNow, you can effortlessly fill out and sign documents online. Leverage the comprehensive tools integrated into this user-friendly and cost-effective platform and transform your method of document management. Whether you need to authorize forms or gather signatures, airSlate SignNow takes care of everything efficiently, with just a few clicks.
Follow this detailed guide:
- Log in to your account or register for a free trial with our service.
- Click +Create to upload a file from your device, cloud storage, or our form repository.
- Access your ‘Quotation Sample A’ in the editor.
- Click Me (Fill Out Now) to finalize the document on your end.
- Add and designate fillable fields for others (if necessary).
- Proceed with the Send Invite settings to solicit eSignatures from others.
- Download, print your copy, or convert it into a reusable template.
No need to worry if you have to collaborate with your colleagues on your Quotation Sample A or send it for notarization—our platform provides you with all the tools necessary to accomplish such tasks. Create an account with airSlate SignNow today and elevate your document management to a new height!
FAQs
-
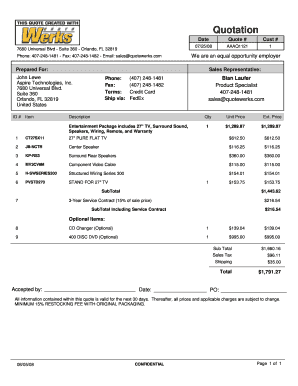
What is a Quotation Sample A and how can it be used?
A Quotation Sample A is a template designed to provide a detailed estimate for products or services. It can be customized to include pricing, descriptions, and terms, making it ideal for businesses looking to streamline their quoting process. With airSlate SignNow, you can easily create and send your Quotation Sample A for electronic signatures, ensuring a professional presentation and quick approval.
-
How can airSlate SignNow help me create a Quotation Sample A?
airSlate SignNow offers user-friendly tools to create a Quotation Sample A effortlessly. You can start with a template or build one from scratch, incorporating your branding and specific details. The platform allows for easy adjustments, ensuring your quotation meets your business needs.
-
What are the pricing options for using airSlate SignNow for my Quotation Sample A?
airSlate SignNow provides flexible pricing plans that cater to businesses of all sizes. Whether you need a basic plan or advanced features for your Quotation Sample A, you can choose a package that fits your budget. Additionally, there’s often a free trial available, allowing you to test the service before committing.
-
Can I integrate airSlate SignNow with other tools for managing my Quotation Sample A?
Yes, airSlate SignNow seamlessly integrates with various business tools, enhancing the management of your Quotation Sample A. Popular integrations include CRM systems, project management tools, and cloud storage services. This connectivity allows for a smoother workflow and better organization.
-
What are the benefits of using airSlate SignNow for my Quotation Sample A?
Using airSlate SignNow for your Quotation Sample A enhances efficiency and professionalism. The electronic signature feature speeds up the approval process, while templates save time in creating quotes. Additionally, the platform ensures your documents are secure and legally compliant.
-
Is it easy to send a Quotation Sample A for eSignature with airSlate SignNow?
Absolutely! Sending a Quotation Sample A for eSignature using airSlate SignNow is straightforward. Simply upload your document, add the recipients' email addresses, and send it off for signing, all within a few clicks.
-
What security measures does airSlate SignNow have for my Quotation Sample A?
airSlate SignNow prioritizes the security of your Quotation Sample A with robust encryption and compliance with industry standards. Your documents are stored securely, and access is controlled, ensuring that sensitive information remains protected throughout the signing process.
Find out other quotation sample a form
- Close deals faster
- Improve productivity
- Delight customers
- Increase revenue
- Save time & money
- Reduce payment cycles